Abstract
Megalin and cubilin, endocytic proteins present in the proximal tubule of the kidney, are responsible for reabsorbing filtered proteins from urine. Our hypothesis was that potential substrates of megalin/cubilin could be identified by examining urinary protein differences between control (WT) mice and kidney-specific megalin knockdown (KD) mice. Using the IonStar proteomics approach, 877 potential megalin/cubilin substrates were discovered, with 23 of these compounds representing known megalin/cubilin substrates. Some of the proteins with the largest fold changes in the urine between KD and WT included the known megalin substrates retinol-binding protein and vitamin D-binding protein. Of the total proteins identified as novel substrates, about three-quarters of compounds had molecular weights (MWs) below 69 kDa, the MW of albumin, and the remaining had higher MWs, with about 5% of the proteins having MWs greater than 150 kDa. Sex differences in the number of identified substrates occurred, but this may be due to differences in kidney megalin expression between both male and female megalin KD and WT animals, with the ratio of megalin between WT and KD being 2.76 and 2.14 for female and male mice, respectively. The top three ingenuity canonical pathways based on the urinary proteins in both female and male KD mice were acute phase response signaling, liver X receptor/retinoid X receptor activation, and intrinsic prothrombin activation pathways. In conclusion, analysis of urine samples from kidney-specific megalin KD and WT mice was found to be useful for the identification of potential endogenous substrates for megalin and cubilin.
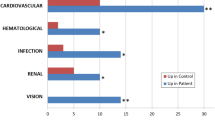
Graphical Abstract








Similar content being viewed by others
Abbreviations
- DBP:
-
Vitamin D binding protein
- FDR:
-
False discovery rate
- FXR:
-
Farnesoid X receptor
- IPA:
-
Ingenuity pathway analysis
- KD:
-
Knockdown
- LC–MS/MS:
-
Liquid chromatography mass spectrometry
- LMW:
-
Low molecular weight
- LXR:
-
Liver X receptor
- MUPs:
-
Major mouse urinary proteins
- MW:
-
Molecular weight
- NPXY:
-
Asn-Pro-any amino acid-tyrosine motifs
- PSM:
-
Peptide-spectrum match
- PT:
-
Proximal tubule
- RBP4:
-
Retinal binding protein
- RXR:
-
Retinoid X receptor
- SPR:
-
Surface plasmon resonance
- WT:
-
Wild type
References
Nielsen R, Christensen EI, Birn H. Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int. 2016;89(1):58–67. https://doi.org/10.1016/j.kint.2015.11.007.
Christensen EI, Birn H. Megalin and cubilin: multifunctional endocytic receptors. Nat Rev Mol Cell Biol. 2002;3(4):258–67. https://doi.org/10.1038/nrm778.
Hammad SM, Barth JL, Knaak C, Argraves WS. Megalin acts in concert with cubilin to mediate endocytosis of high density lipoproteins. J Biol Chem. 2000;275(16):12003–8. https://doi.org/10.1074/jbc.275.16.12003.
Christensen EI, Moskaug JO, Vorum H, Jacobsen C, Gundersen TE, Nykjaer A, et al. Evidence for an essential role of megalin in transepithelial transport of retinol. J Am Soc Nephrol. 1999;10(4):685–95.
Gonzalez-Villalobos R, Klassen RB, Allen PL, Navar LG, Hammond TG. Megalin binds and internalizes angiotensin II. Am J Physiol Renal Physiol. 2005;288(2):F420–7. https://doi.org/10.1152/ajprenal.00243.2004.
Gonzalez-Villalobos R, Klassen RB, Allen PL, Johanson K, Baker CB, Kobori H, et al. Megalin binds and internalizes angiotensin-(1–7). Am J Physiol Renal Physiol. 2006;290(5):F1270–5. https://doi.org/10.1152/ajprenal.00164.2005.
Nykjaer A, Dragun D, Walther D, Vorum H, Jacobsen C, Herz J, et al. An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell. 1999;96(4):507–15.
Leheste JR, Rolinski B, Vorum H, Hilpert J, Nykjaer A, Jacobsen C, et al. Megalin knockout mice as an animal model of low molecular weight proteinuria. Am J Pathol. 1999;155(4):1361–70. https://doi.org/10.1016/S0002-9440(10)65238-8.
Orlando RA, Rader K, Authier F, Yamazaki H, Posner BI, Bergeron JJ, et al. Megalin is an endocytic receptor for insulin. J Am Soc Nephrol. 1998;9(10):1759–66.
Gburek J, Birn H, Verroust PJ, Goj B, Jacobsen C, Moestrup SK, et al. Renal uptake of myoglobin is mediated by the endocytic receptors megalin and cubilin. J Am Soc Nephrol. 2002;13:486a-a487.
Moestrup SK, Birn H, Fischer PB, Petersen CM, Verroust PJ, Sim RB, et al. Megalin-mediated endocytosis of transcobalamin-vitamin-B12 complexes suggests a role of the receptor in vitamin-B12 homeostasis. Proc Natl Acad Sci U S A. 1996;93(16):8612–7.
Sousa MM, Norden AG, Jacobsen C, Willnow TE, Christensen EI, Thakker RV, et al. Evidence for the role of megalin in renal uptake of transthyretin. J Biol Chem. 2000;275(49):38176–81. https://doi.org/10.1074/jbc.M002886200.
Nykjaer A, Fyfe JC, Kozyraki R, Leheste J-R, Jacobsen C, Nielsen MS, et al. Cubilin dysfunction causes abnormal metabolism of the steroid hormone 25(OH) vitamin D. PNAS. 2001;98(24):13895–900. https://doi.org/10.1073/pnas.241516998.
Liu CP, Hu Y, Lin JC, Fu HL, Lim LY, Yuan ZX. Targeting strategies for drug delivery to the kidney: From renal glomeruli to tubules. Med Res Rev. 2019;39(2):561–78. https://doi.org/10.1002/med.21532.
Lameire N. Nephrotoxicity of recent anti-cancer agents. Clin Kidney J. 2014;7(1):11–22. https://doi.org/10.1093/ckj/sft135.
Hori Y, Aoki N, Kuwahara S, Hosojima M, Kaseda R, Goto S, et al. Megalin blockade with cilastatin suppresses drug-induced nephrotoxicity. J Am Soc Nephrol. 2017;28(6):1783–91. https://doi.org/10.1681/asn.2016060606.
Bryniarski MA, Zhao B, Chaves LD, Mikkelsen JH, Yee BM, Yacoub R, et al. Immunoglobulin G is a novel substrate for the endocytic protein megalin. AAPS J. 2021;23(2):40. https://doi.org/10.1208/s12248-021-00557-1.
Leheste J-R, Rolinski B, Vorum H, Hilpert J, Nykjaer A, Jacobsen C, et al. Megalin Knockout mice as an animal model of low molecular weight proteinuria. Am J Pathol. 1999;155(4):1361–70. https://doi.org/10.1016/S0002-9440(10)65238-8.
Shen S, An B, Wang X, Hilchey SP, Li J, Cao J, et al. Surfactant cocktail-aided extraction/precipitation/on-pellet digestion strategy enables efficient and reproducible sample preparation for large-scale quantitative proteomics. Anal Chem. 2018;90(17):10350–9. https://doi.org/10.1021/acs.analchem.8b02172.
Tu C, Mammen MJ, Li J, Shen X, Jiang X, Hu Q, et al. Large-scale, ion-current-based proteomics investigation of bronchoalveolar lavage fluid in chronic obstructive pulmonary disease patients. J Proteome Res. 2014;13:627.
Tu C, Fiandalo MV, Pop E, Stocking JJ, Azabdaftari G, Li J, et al. Proteomic analysis of charcoal-stripped fetal bovine serum reveals changes in the insulin-like growth factor signaling pathway. J Proteome Res. 2018;17(9):2963–77. https://doi.org/10.1021/acs.jproteome.8b00135.
Tu C, Li J, Young R, Page BJ, Engler F, Halfon MS, et al. Combinatorial peptide ligand library treatment followed by a dual-enzyme, dual-activation approach on a nanoflow liquid chromatography/orbitrap/electron transfer dissociation system for comprehensive analysis of swine plasma proteome. Anal Chem. 2011;83(12):4802–13. https://doi.org/10.1021/ac200376m.
Tu C, Bu Y, Vujcic M, Shen S, Li J, Qu M, et al. Ion current-based proteomic profiling for understanding the inhibitory effect of tumor necrosis factor alpha on myogenic differentiation. J Proteome Res. 2016;15(9):3147–57. https://doi.org/10.1021/acs.jproteome.6b00321.
Shen X, Shen S, Li J, Hu Q, Nie L, Tu C, et al. An IonStar experimental strategy for MS1 ion current-based quantification using ultrahigh-field orbitrap: reproducible, in-depth, and accurate protein measurement in large cohorts. J Proteome Res. 2017;16(7):2445–56. https://doi.org/10.1021/acs.jproteome.7b00061.
Shen X, Shen S, Li J, Hu Q, Nie L, Tu C, et al. IonStar enables high-precision, low-missing-data proteomics quantification in large biological cohorts. PNAS. 2018;115(21):E4767–76. https://doi.org/10.1073/pnas.1800541115.
Wang X, Jin L, Hu C, Shen S, Qian S, Ma M, et al. Ultra-high-resolution IonStar strategy enhancing accuracy and precision of MS1-based proteomics and an extensive comparison with state-of-the-art SWATH-MS in large-cohort quantification. Anal Chem. 2021;93(11):4884–93. https://doi.org/10.1021/acs.analchem.0c05002.
Sadygov RG, Maroto FM, Huhmer AF. ChromAlign: a two-step algorithmic procedure for time alignment of three-dimensional LC-MS chromatographic surfaces. Anal Chem. 2006;78(24):8207–17. https://doi.org/10.1021/ac060923y.
Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2013;30(4):523–30. https://doi.org/10.1093/bioinformatics/btt703.
Koral K, Li H, Ganesh N, Birnbaum MJ, Hallows KR, Erkan E. Akt recruits Dab2 to albumin endocytosis in the proximal tubule. Am J Physiol Renal Physiol. 2014;307(12):F1380–9. https://doi.org/10.1152/ajprenal.00454.2014.
Nagai J, Christensen EI, Morris SM, Willnow TE, Cooper JA, Nielsen R. Mutually dependent localization of megalin and Dab2 in the renal proximal tubule. American Journal of Physiology-Renal Physiology. 2005;289(3):F569–76. https://doi.org/10.1152/ajprenal.00292.2004.
Gburek J, Verroust PJ, Willnow TE, Fyfe JC, Nowacki W, Jacobsen C, et al. Megalin and cubilin are endocytic receptors involved in renal clearance of hemoglobin. J Am Soc Nephrol. 2002;13(2):423–30.
Oyama Y, Takeda T, Hama H, Tanuma A, Iino N, Sato K, et al. Evidence for megalin-mediated proximal tubular uptake of L-FABP, a carrier of potentially nephrotoxic molecules. Lab Invest. 2005;85(4):522–31. https://doi.org/10.1038/labinvest.3700240.
Birn H, Vorum H, Verroust PJ, Moestrup SK, Christensen EI. Receptor- associated protein is important for normal processing of megalin in kidney proximal tubules. J Am Soc Nephrol. 2000;11(2):191–202.
Kounnas MZ, Loukinova EB, Stefansson S, Harmony JAK, Brewer BH, Strickland DK, et al. Identification of glycoprotein-330 as an endocytic receptor for apolipoprotein-J/clusterin. J Biol Chem. 1995;270(22):13070–5. https://doi.org/10.1074/jbc.270.22.13070.
Godyna S, Liau G, Popa I, Stefansson S, Argraves WS. Identification of the low-density- lipoprotein receptor-related protein (Lrp) as an endocytic receptor for thrombospondin-1. J Cell Biol. 1995;129(5):1403–10. https://doi.org/10.1083/jcb.129.5.1403.
Hama H, Saito A, Takeda T, Tanuma A, Xie Y, Sato K, et al. Evidence indicating that renal tubular metabolism of leptin is mediated by megalin but not by the leptin receptors. Endocrinology. 2004;145(8):3935–40. https://doi.org/10.1210/en.2004-0074.
Nielsen R, Courtoy PJ, Jacobsen C, Dom G, Lima WR, Jadot M, et al. Endocytosis provides a major alternative pathway for lysosomal biogenesis in kidney proximal tubular cells. Proc Natl Acad Sci U S A. 2007;104(13):5407–12. https://doi.org/10.1073/pnas.0700330104.
Kaseda R, Iino N, Hosojima M, Takeda T, Hosaka K, Kobayashi A, et al. Megalin-mediated endocytosis of cystatin C in proximal tubule cells. Biochem Biophys Res Commun. 2007;357(4):1130–4. https://doi.org/10.1016/j.bbrc.2007.04.072.
Kounnas MZ, Chappell DA, Strickland DK, Argraves WS. Glycoprotein-330, a member of the low-density-lipoprotein receptor family, binds lipoprotein-lipase in-vitro. J Biol Chem. 1993;268(19):14176–81.
Willnow TE, Goldstein JL, Orth K, Brown MS, Herz J. Low-density-lipoprotein receptor- related protein and Gp330 bind similar ligands, including plasminogen activator-inhibitor complexes and lactoferrin, an inhibitor of chylomicron remnant clearance. J Biol Chem. 1992;267(36):26172–80.
Kanalas JJ. Analysis of plasmin binding and urokinase activation of plasminogen bound to the Heymann nephritis autoantigen, Gp330. Arch Biochem Biophys. 1992;299(2):255–60. https://doi.org/10.1016/0003-9861(92)90272-X.
Kanalas JJ, Makker SP. Identification of the rat Heymann nephritis autoantigen (GP330) as a receptor site for plasminogen. J Biol Chem. 1991;266(17):10825–9.
Cui S, Verroust PJ, Moestrup SK, Christensen EI. Megalin/gp330 mediates uptake of albumin in renal proximal tubule. Am J Physiol. 1996;271(4 Pt 2):F900–7. https://doi.org/10.1152/ajprenal.1996.271.4.F900.
Birn H, Zhai X, Holm J, Hansen SI, Jacobsen C, Christensen EI, et al. Megalin binds and mediates cellular internalization of folate binding protein. FEBS J. 2005;272(17):4423–30. https://doi.org/10.1111/j.1742-4658.2005.04857.x.
Faber K, Hvidberg V, Moestrup SK, Dahlback B, Nielsen LB. Megalin is a receptor for apolipoprotein M, and kidney-specific megalin-deficiency confers urinary excretion of apolipoprotein M. Mol Endocrinol. 2006;20(1):212–8. https://doi.org/10.1210/me.2005-0209.
Sun Y, Lu X, Danser AHJ. Megalin: a novel determinant of renin-angiotensin system activity in the kidney? Curr Hypertens Rep. 2020;22(4):30. https://doi.org/10.1007/s11906-020-01037-1.
Kukida M, Sawada H, Daugherty A, Lu HS. Megalin: a bridge connecting kidney, the renin- angiotensin system, and atherosclerosis. Pharmacol Res. 2020;151: 104537. https://doi.org/10.1016/j.phrs.2019.104537.
Geertzen HGM, Ouderaa FJGvd, Kassenaar AAH. Isolation and metabolism of male sex- dependent urinary protein from rats. Acta Endocrinol (Copenh). 1973;72(1):197. doi: https://doi.org/10.1530/acta.0.0720197.
Lane SE, Neuhaus OW. Multiple forms of 2 u, a sex-dependent urinary protein of the adult male rat. Biochim Biophys Acta. 1972;263(2):433–40. https://doi.org/10.1016/0005-2795(72)90095-5.
Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318(Pt 1):1–14. https://doi.org/10.1042/bj3180001.
Baylis C. Sexual dimorphism, the aging kidney, and involvement of nitric oxide deficiency. Semin Nephrol. 2009;29(6):569–78. https://doi.org/10.1016/j.semnephrol.2009.07.003.
Munger K, Baylis C. Sex differences in renal hemodynamics in rats. Am J Physiol. 1988;254(2 Pt 2):F223–31. https://doi.org/10.1152/ajprenal.1988.254.2.F223.
Remuzzi A, Puntorieri S, Mazzoleni A, Remuzzi G. Sex related differences in glomerular ultrafiltration and proteinuria in Munich-Wistar rats. Kidney Int. 1988;34(4):481–6. https://doi.org/10.1038/ki.1988.206.
Dickinson H, Moritz KM, Kett MM. A comparative study of renal function in male and female spiny mice - sex specific responses to a high salt challenge. Biol Sex Differ. 2013;4(1):21. https://doi.org/10.1186/2042-6410-4-21.
BaireyMerz CN, Dember LM, Ingelfinger JR, Vinson A, Neugarten J, Sandberg KL, et al. Sex and the kidneys: current understanding and research opportunities. Nat Rev Nephrol. 2019;15(12):776–83. https://doi.org/10.1038/s41581-019-0208-6.
Yang H, Zhang W, Lu S, Lu G, Zhang H, Zhuang Y, et al. Mup-knockout mice generated through CRISPR/Cas9-mediated deletion for use in urinary protein analysis. Acta Biochim Biophys Sin (Shanghai). 2016;48(5):468–73. https://doi.org/10.1093/abbs/gmw003.
Leheste JR, Melsen F, Wellner M, Jansen P, Schlichting U, Renner-Muller I, et al. Hypocalcemia and osteopathy in mice with kidney-specific megalin gene defect. FASEB J. 2003;17(2):247–9. https://doi.org/10.1096/fj.02-0578fje.
Bryniarski MA, Zhao B, Chaves LD, Mikkelsen JH, Yee BM, Yacoub R, et al. Immunoglobulin G is a novel substrate for the endocytic protein megalin. Aaps j. 2021;23(2):40. https://doi.org/10.1208/s12248-021-00557-1.
Funding
Financial support by the Center of Protein Therapeutics, University at Buffalo (MEM).
Author information
Authors and Affiliations
Contributions
Participated in research design: Morris, Qu, and Zhao.
Conducted experiments: Morris, Zhao, Tu, and Shen.
Contributed new reagents or analytic tools: Morris and Qu.
Performed data analysis: Morris, Tu, Shen, and Zhao.
Wrote or contributed to the writing of the manuscript: Morris, Qu, Tu, Shen, and Zhao.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, B., Tu, C., Shen, S. et al. Identification of Potential Megalin/Cubilin Substrates Using Extensive Proteomics Quantification from Kidney Megalin-Knockdown Mice. AAPS J 24, 109 (2022). https://doi.org/10.1208/s12248-022-00758-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-022-00758-2